List of current projects:
- DNA Synthesis Automation and Technology Development
- Drug Discovery and Generating Impact
- Synthetic Genomics
- YeastFab
DNA Synthesis Automation and Technology Development
DNA Synthesis Automation
To close the gap between design and fabrication in synthetic biology, we are proposing to automate the DNA synthesis process, utilizing liquid handling robots and other laboratory automation equipment. The first goal is to implement basic molecular biology operations on a liquid handler, such as cherry picking, PCR, gel electrophoresis, cloning, bacterial and yeast transformation, plasmid DNA miniprep, and colony screening. In each step of molecular biology experiments, there will be failures, and how to reschedule these failures in an optimized fashion is a bigger challenge. Therefore, the second goal is to develop a workflow management system which schedules experiments based on given optimization objectives, such as cost, total time and minimal hands-on time.
EMMA
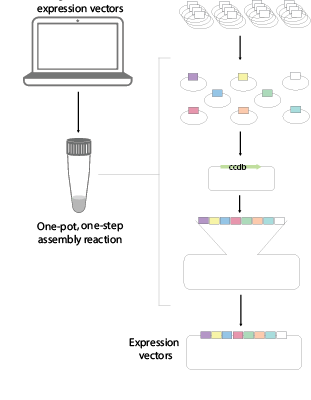
Traditional cloning methods often require laborious manual design and assembly of plasmids using tailored sequential cloning steps. This process can be protracted, complicated, expensive and error-prone. In our lab, we have developed an extensible mammalian modular assembly kit (EMMA), that facilitate the efficient design and production of bespoke vectors relieving a current bottleneck for researchers. The method supports assembly of combinatorial libraries, and hierarchical assembly for production of larger multigenetic cargos. We are also investigating new efficient ways of delivering megabase-size DNA constructs into mammalian cells. EMMA is compatible with automated production and provides new opportunities for mammalian synthetic biology.
DNA Recombinase toolkit for synthetic biology
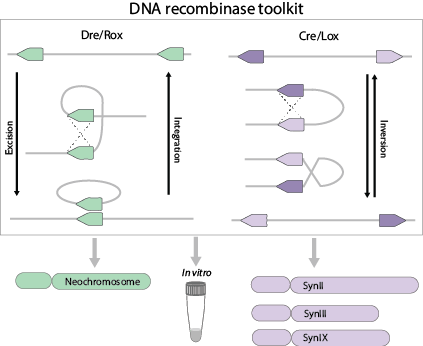
Site-specific DNA recombinases are a family of DNA enzymes that can recognize specific DNA sequences and drive DNA recombination between specific sites to achieve deletion, inversion, integration or transposition of target DNA fragments. The Sc2.0 synthetic yeast genome is designed to contain numerous LoxP sites located throughout, enabling random genome rearrangement for accelerated yeast evolution, a process called SCRaMbLE-(Synthetic Chromosome Recombination and Modification by LoxP-mediated Evolution).
Our group is also constructing a tRNA Neochromosome that contains all tRNA genes relocated from the synthetic yeast genome. For the purpose of SCRaMbLE, a recombination system based on Dre/rox achieves orthogonal evolution of the neochromosome.
Drug Discovery and Generating Impact
Synthetic Natural Products (SynNP)
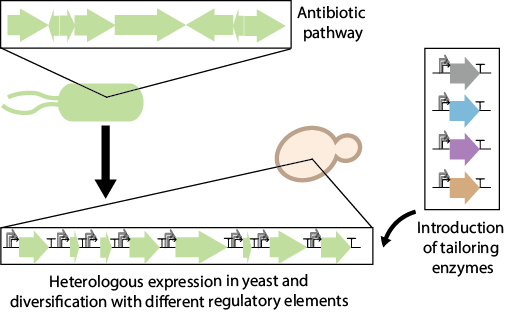
The SynNP project aims to apply synthetic biology to the discovery and design of new antibiotics against tuberculosis and other infectious diseases. Based on a large Bio Synthetic Gene (BSG) library and yeast promoter library (YeastFab), we are able to mix and match BSGs to assemble diversified synthetic pathway circuits, which can produce chemical libraries for the treatment of tuberculosis infection. With further high-throughput screening, we aim to discover new treatments and improve the yield of medicines against tuberculosis.
Synthetic Genomics
Sc 2.0
Our group is heavily involved with the synthetic yeast genome consortium (Sc2.0, www.syntheticyeast.org), which aims to re-design and synthesize a 12 Mb designer yeast genome de novo (for details see Dymond, Nature 2010). Our group will design and synthesize at least one chromosome in house, so the success of this project will be of great significance and highly valuable to the field of synthetic biology.
Evolution of Synthetic Yeast genomes (IESY)
The Induced Evolution of Synthetic Yeast genomes (IESY) project builds on pioneering developments from the Saccharomyces cerevisiae 2.0 (Sc2.0) consortium. IESY represents an innovative concept in synthetic biology to explore combinatory diversity in pre-programmed synthetic genomes to evolve new and useful functions. Using the system for Synthetic Chromosome Rearrangement and Modification by LoxP-mediated evolution (SCRaMbLE) we are able to generate high-fitness yeast cells with synthetic chromosomes under specific conditions. The SCRaMbLE system can generate genome diversity with the capability of genome minimization and the ability to produce new biological functions through rapid evolution of genome content, copy number and gene order.
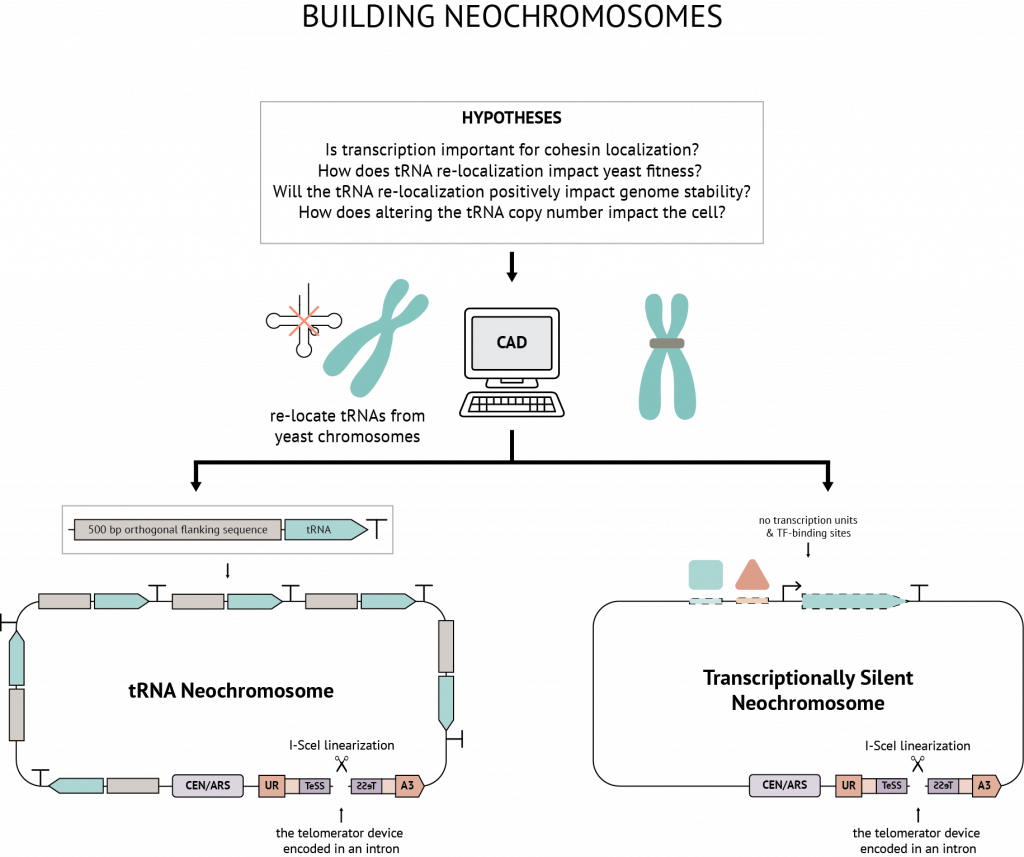
tRNA Neochromosome
This project is part of the international synthetic yeast consortium. The goal of this project is to design and build a tRNA neochromosome in yeast, hosting all of the tRNAs which will be removed from the synthetic yeast genome.
tRNA genes have been shown to be hotspots for DNA instability for several reasons. First, tRNA genes are often the target of 5’ upstream retrotransposon incorporation. Second, tRNA genes are transcribed heavily, which can lead to DNA damage caused by collisions between the replication and transcriptional machineries. Furthermore, retrotransposon insertions produce highly-homologous repeats leading to homologous recombination between chromosomal regions.
Thus, as part of Sc2.0, all tRNA genes will be moved to their own dedicated chromosome (a “Party Chromosome”). All introns will also be removed from tDNA, as another goal of the project is to observe the effect of a yeast genome with fewer introns. rox Recombination sites will be placed between the tRNA genes, which will enable us ‘SCRaMbLE’ the neochromosome and observe whether there is a preference for tRNA copy number under different conditions.
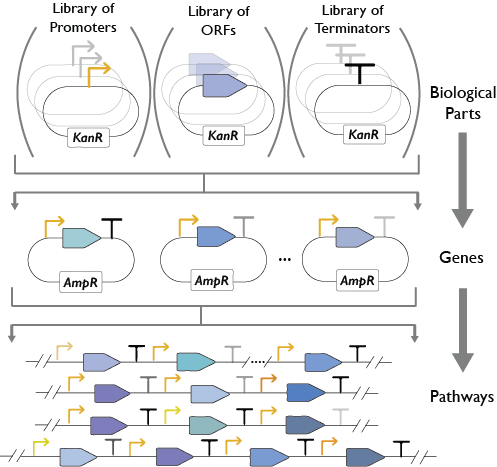
YeastFab
The YeastFab project aims to construct the first comprehensive library of standardised and well-characterised biological parts (promoters, open-reading frames and terminators) for the Saccharomyces cerevisiae chasis. Our group along with researchers at the Dai laboratory at Tsinghua university (China) are working to domesticate over 6,000 promoters, ORFs and terminators to the YeastFab standard using high throughput biology and automation. The promoters and terminators are subsequently characterised by flow cytometry to determine their activity both in normal and stress conditions including; oxidative stress (H2O2), heat (37℃) and nutrient starvation (in medium lacking glucose, nitrogen starvation or in water). We have also devised a golden gate-based YeastFab assembly strategy that enables the rapid and cost-effective assembly of pathways. Not only does this enable assembly of pathways within a week but it is also scalable and enables combinatorial assembly of parts. We expect that through standardisation of parts, this will reduce the time and cost of yeast metabolic engineering thus accelerating the iterations of the Design-Build-Test cycle for heterologous expression of pathways.